Modules
-
Module 1: Data Exploration - Biomarkers for cerebral Malaria in protein expression data
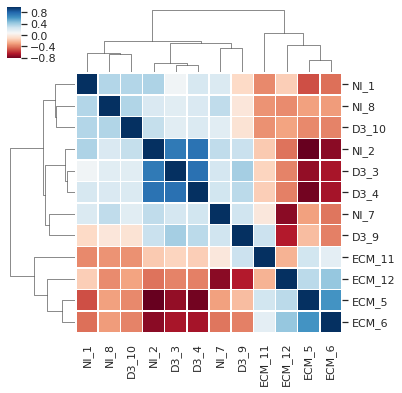
This module covers dataset I/O handling, modelling and normalisation in Pandas, principal component analysis and differential gene expression analysis. The case data is on quantification of relative protein levels in malaria-infected mice from work by Tiberti, N et al Scientific Reports 2016.
-
Module 2: Data Visualisation - Sequence features for thermostability in proteins from extremophiles

View this notebook This module covers efficient data processing in Numpy, analysis and visualisation of biological sequences and graphing in Seaborn and Matplotlib. The case data comes from a dataset of 7.7 million bacterial sequences with associated temperature data compiled by the iGEM Potsdam team for Kaggle, collected from the Bacterial Diversity Metadatabase and UniProt.
-
Module 4: Machine-learning classification of benign and malignant tumors
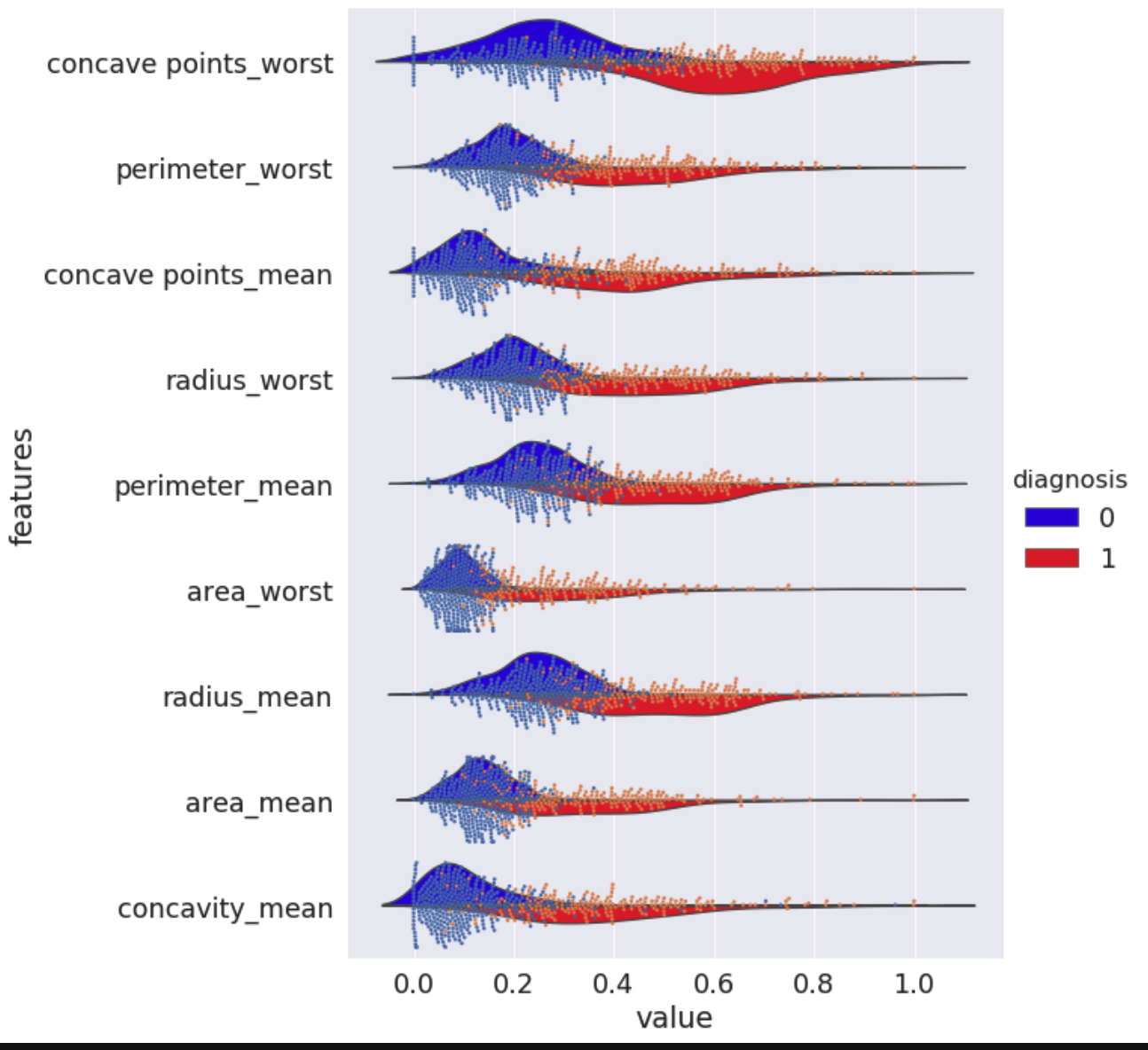
In this module we will guide you through preprocessing and design of multiple machine-learning prediction models based on imaging features of tumor fine needle aspirates. We will perform exploratory data analysis of the clinical dataset, cover key steps and decisions in preprocessing, undergo comprehensive comparison of different machine learning architectures and finally evaluate all our trained models. This module, like all our modules, focuses on visual understanding of decisions we make and use of workflows that will apply to any dataset.
-
Module 5: Protein Sequences Analysis - Analysing antibodies binding SARS-CoV-2
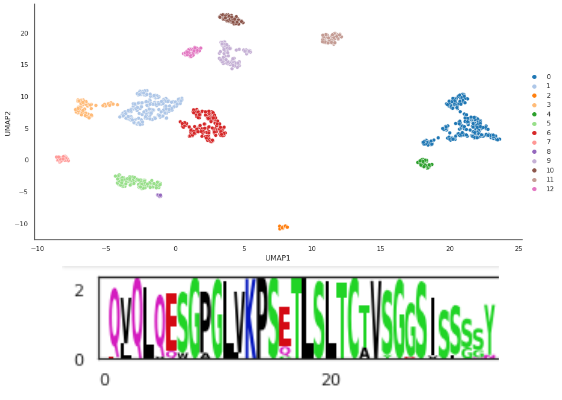
This module covers different analytical methods of protein sequences such as sequence alignments, clustering and logoplots. We will be looking at coronavirus binding antibodies, exploring how they can be clustered using PCA, t-SNE and UMAP, how these clusters can be visualized using logoplots and how to interpret these findings.
